Richard Grewelle, PhD
Researching
Ocean health depends on our understanding of the intimate iteractions between gene, organism, and ecosystem.
Research
The impact humans have on land is evident, and as we extend our reach towards the ocean, its health is jeopardized by the change we create. Ecosystems are exposed to new stressors, including exploitation and disease. To protect fragile species, I use genetics and mathematics to understand the defensive and destructive role of innate immunity in the face of a changing pathogenic seascape. Genes controlling immunity also may regulate symbiosis in coral and anemones, which are now bleaching at unprecedented rates. We now have the opportunity to interrogate these genes with modern technology, which will deliver insights into the mechanisms of symbiosis and pathogenesis in corals, ultimately guiding us towards solutions to these threats to ocean health.
- Current Position: Post-doctoral Scholar, Department of Genetics, Stanford University
- Faculty Advisor: John Pringle
Publications
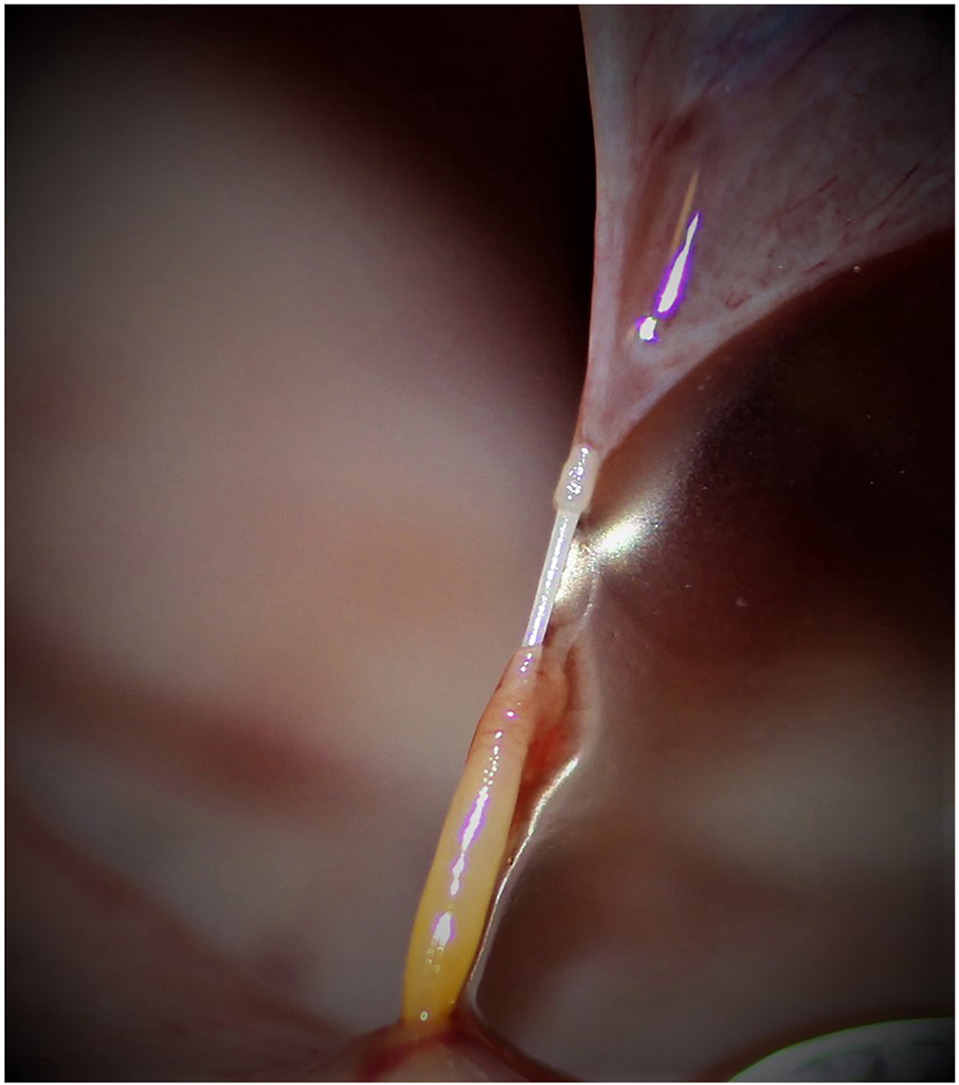
Abstract: Acanthocephalan peritonitis (AP; trans-intestinal migration of acanthocephalan parasites into the peritoneal cavity resulting in severe peritonitis), is a common cause of mortality in southern sea otters (Enhydra lutris nereis). Although Profilicollis spp. acanthocephalans have been implicated in these infections, the species causing AP has been an important unresolved question for decades. We used morphological and molecular techniques to characterize acanthocephalans from the gastrointestinal (GI) tract and peritoneal omentum of eighty necropsied southern sea otters. Only P. altmani was found to have perforated through the intestinal wall and migrated into the peritoneal cavity of examined sea otters, resulting in AP. Morphological and molecular criteria confirmed that Profilicollis kenti was synonymous with P. altmani. A second Profilicollis sp., likely P. botulus, was present only in the intestinal lumen, did not penetrate through the intestinal wall, and was not associated with AP.

Abstract: Multiple stressors to species and ecosystems are pervasive and escalating. Effective management and mitigation of these pressures requires ecological risk assessment (ERA), but data are often lacking for detailed, quantitative risk assessment. Data-poor ERAs have been developed and widely applied to terrestrial, marine, and freshwater ecosystems. Current frameworks, such as the Productivity-Susceptibility Analysis (PSA), are limited to single stressors and were not developed on statistical grounds. Previous work has partly addressed these limitations by incorporating multiple stressors (e.g. Aggregated Susceptibility) and a statistical basis (rPSA). However, the more robust rPSA is more difficult to implement than the PSA. To overcome this barrier, here we develop EcoRAMS (Ecological Risk Assessment of Multiple Stressors), which provides statistically-robust ecological risk assessments of multiple stressors in data-poor contexts. The web app format of EcoRAMS.net lowers the barrier of use for practitioners and scientists at any level of statistical training.

Abstract: CRISPR gene drives could revolutionize the control of infectious diseases by accelerating the spread of engineered traits that limit parasite transmission in wild populations. Gene drive technology in mollusks has received little attention despite the role of freshwater snails as hosts of parasitic flukes causing 200 million annual cases of schistosomiasis. A successful drive in snails must overcome self-fertilization, a common feature of host snails which could prevents a drive’s spread. Here we developed a novel population genetic model accounting for snails’ mixed mating and population dynamics, susceptibility to parasite infection regulated by multiple alleles, fitness differences between genotypes, and a range of drive characteristics. We integrated this model with an epidemiological model of schistosomiasis transmission to show that a snail population modification drive targeting immunity to infection can be hindered by a variety of biological and ecological factors; yet under a range of conditions, disease reduction achieved by chemotherapy treatment of the human population can be maintained with a drive. Alone a drive modifying snail immunity could achieve significant disease reduction in humans several years after release. These results indicate that gene drives, in coordination with existing public health measures, may become a useful tool to reduce schistosomiasis burden in selected transmission settings with effective CRISPR construct design and evaluation of the genetic and ecological landscape.

Abstract: The productivity susceptibility analysis (PSA) is a widely used method to rapidly assess species risk to fishing activities in data-poor fisheries. A step in ecological risk assessments and used in data-poor assessment for sustainable fisheries certification programmes (e.g. MSC) and recommendation lists (e.g. Seafood Watch), the PSA is semi-quantitative, yet little attention has been given to the theoretical basis of this analysis. Current thresholds designating low-, medium- and high-risk categories divide the PSA plot by equal area, assuming area corresponds to likelihood. We show that plot area does not correspond to likelihood, however, and existing thresholds need revision due to the non-uniform distribution of vulnerability scores on the PSA plot. The probability of medium risk assignment increases with the number of attributes used to characterize productivity and susceptibility. Here, we present a novel and statistically robust method to derive vulnerability, where threshold values between the risk categories are adjusted with the number of attributes used in the assessment. Our comprehensive framework accounts for all variations in the method, including logarithmic scaling of axes, weighting of attributes and scoring procedures. Simulated results across a range of conditions and comparative evaluation of 302 species in five studies show that one-third of species may be re-categorized with the new PSA approach. Importantly, the existing PSA approach underestimates risk by up to 35% when compared with the new method. These findings have strong implications for management of data-poor fisheries. We recommend adoption of this approach to the PSA to better resolve species’ risk.

Abstract: COVID-19 has become a global pandemic, resulting in nearly three hundred thousand deaths distributed heterogeneously across countries. Estimating the infection fatality rate (IFR) has been elusive due to the presence of asymptomatic or mildly symptomatic infections and lack of testing capacity. We analyze global data to derive the IFR of COVID-19. Estimates of COVID-19 IFR in each country or locality differ due to variable sampling regimes, demographics, and healthcare resources. We present a novel statistical approach based on sampling effort and the reported case fatality rate of each country. The asymptote of this function gives the global IFR. Applying this asymptotic estimator to cumulative COVID-19 data from 139 countries reveals a global IFR of 1.04% (CI: 0.77%,1.38%). Deviation of countries’ reported CFR from the estimator does not correlate with demography or per capita GDP, suggesting variation is due to differing testing regimes or reporting guidelines by country. Estimates of IFR through seroprevalence studies and point estimates from case studies or sub-sampled populations are limited by sample coverage and cannot inform a global IFR, as mortality is known to vary dramatically by age and treatment availability. Our estimated IFR aligns with many previous estimates and is the first attempt at a global estimate of COVID-19 IFR.

Abstract: The first signs of sea star wasting disease (SSWD) epidemic occurred in just few months in 2013 along the entire North American Pacific coast. Disease dynamics did not manifest as the typical travelling wave of reaction-diffusion epidemiological model, suggesting that other environmental factors might have played some role. To help explore how external factors might trigger disease, we built a coupled oceanographic-epidemiological model and contrasted three hypotheses on the influence of temperature on disease transmission and pathogenicity. Models that linked mortality to sea surface temperature gave patterns more consistent with observed data on sea star wasting disease, which suggests that environmental stress could explain why some marine diseases seem to spread so fast and have region-wide impacts on host populations.

Abstract: Emergence of new viral diseases is linked to mutation or recombination events. The likelihood of cross-species transmission is related to phenotypic plasticity of a virus and its capacity to produce genetically variable progeny. Herein a model is described connecting the production of genetic variability with increasing genome size. Comparing all known zoonotic viral genome sizes to known non-zoonotic viral genome sizes demonstrates that zoonotic viruses have significantly larger genomes. These results support the notion that large viral genomes are important in producing new zoonotic disease, and suggest that genome size may be a useful surrogate in screening for potential zoonotic viruses.

Abstract: Schistosomiasis is one of the most important and widespread neglected tropical diseases (NTD), with over 200 million people infected in more than 70 countries; the disease has nearly 800 million people at risk in endemic areas. Although mass drug administration is a cost-effective approach to reduce occurrence, extent, and severity of the disease, it does not provide protection to subsequent reinfection. Interventions that target the parasites’ intermediate snail hosts are a crucial part of the integrated strategy required to move toward disease elimination. The recent revolution in gene drive technology naturally leads to questions about whether gene drives could be used to efficiently spread schistosome resistance traits in a population of snails and whether gene drives have the potential to contribute to reduced disease transmission in the long run. Responsible implementation of gene drives will require solutions to complex challenges spanning multiple disciplines, from biology to policy. This Review Article presents collected perspectives from practitioners of global health, genome engineering, epidemiology, and snail/schistosome biology and outlines strategies for responsible gene drive technology development, impact measurements of gene drives for schistosomiasis control, and gene drive governance. Success in this arena is a function of many factors, including gene-editing specificity and efficiency, the level of resistance conferred by the gene drive, how fast gene drives may spread in a metapopulation over a complex landscape, ecological sustainability, social equity, and, ultimately, the reduction of infection prevalence in humans. With combined efforts from across the broad global health community, gene drives for schistosomiasis control could fortify our defenses against this devastating disease in the future.

Abstract: Perhaps the most important recent advance in species delimitation has been the development of model-based approaches to objectively diagnose species diversity from genetic data. Additionally, the growing accessibility of next-generation sequence data sets provides powerful insights into genome-wide patterns of divergence during speciation. However, applying complex models to large data sets is time-consuming and computationally costly, requiring careful consideration of the influence of both individual and population sampling, as well as the number and informativeness of loci on species delimitation conclusions. Here, we investigated how locus number and information content affect species delimitation results for an endangered Mexican salamander species, Ambystoma ordinarium. We compared results for an eight-locus, 137-individual data set and an 89-locus, seven-individual data set. For both data sets, we used species discovery methods to define delimitation models and species validation methods to rigorously test these hypotheses. We also used integrated demographic model selection tools to choose among delimitation models, while accounting for gene flow. Our results indicate that while cryptic lineages may be delimited with relatively few loci, sampling larger numbers of loci may be required to ensure that enough informative loci are available to accurately identify and validate shallow-scale divergences. These analyses highlight the importance of striking a balance between dense sampling of loci and individuals, particularly in shallowly diverged lineages. They also suggest the presence of a currently unrecognized, endangered species in the western part of A. ordinarium's range.
Teaching
2017
Introductory course on biological analysis and inquiry with case studies on human and animal infectious diseases.
30 undergraduates in twice weekly lecture, split into 4 weekly, TA-led recitation sections.
Course instructors: Mary Beth Mudgett, Martha Cyert, Hunter Fraser, Waheeda Khalfan
2018
Lecture, discussion, and seminar course on contemporary topics of marine conservation.
10 undergraduate and 5 graduate students in weekly 6 hour hybrid format class.
Course instructor: Larry Crowder
2021 & 2022
Lecture, discussion, and stats lab course encompassing modeling in population dynamics, species distributions, fisheries, and population genetics.
6 undergraduate and 4 graduate students in twice weekly hybrid format class.
Course instructors: Giulio De Leo, Maurice Goodman, Gianalberto Losapio, Richard Grewelle
Education
Hopkins Marine Station
Degree: Ph.D. in Biology
Year: 2022
- Stanford Graduate Fellow
- ARCS Foundation Scholar
- Excellence in Teaching Award
- Samuel Karlin Prize in Mathematical Biology (Given to the graduate student whose dissertation reaches the highest standard of Mathematical Biology, this award honors Professor Samuel Karlin of Stanford's Department of Mathematics.)
Awards:
Degrees: B.S. Hons Biology, B.S. Hons Chemistry, B.S. Hons Mathematics
Year: 2016
Awards: